DICOM
Encord provides native support for Digital Imaging and Communications in Medicine (DICOM) browser rendering and data annotations. With the DICOM editor, you can:
- Annotate modalities such as CT, X-ray, and MRI
- Label using any annotation type in 2D (with 3D in the works), and seamlessly toggle between axial, coronal, and sagittal views
- Render 20,000 pixel intensities and set custom window widths & levels
- Natively display DICOM metadata
- Track and interpolate objects between slices
- Reduce manual annotations with automation features
We recommend using the Firefox browser when working with DICOM files, as it doesn't have a tab memory limit. Tab memory limits in other browsers such as Chrome may cause issues when loading large DICOM files.
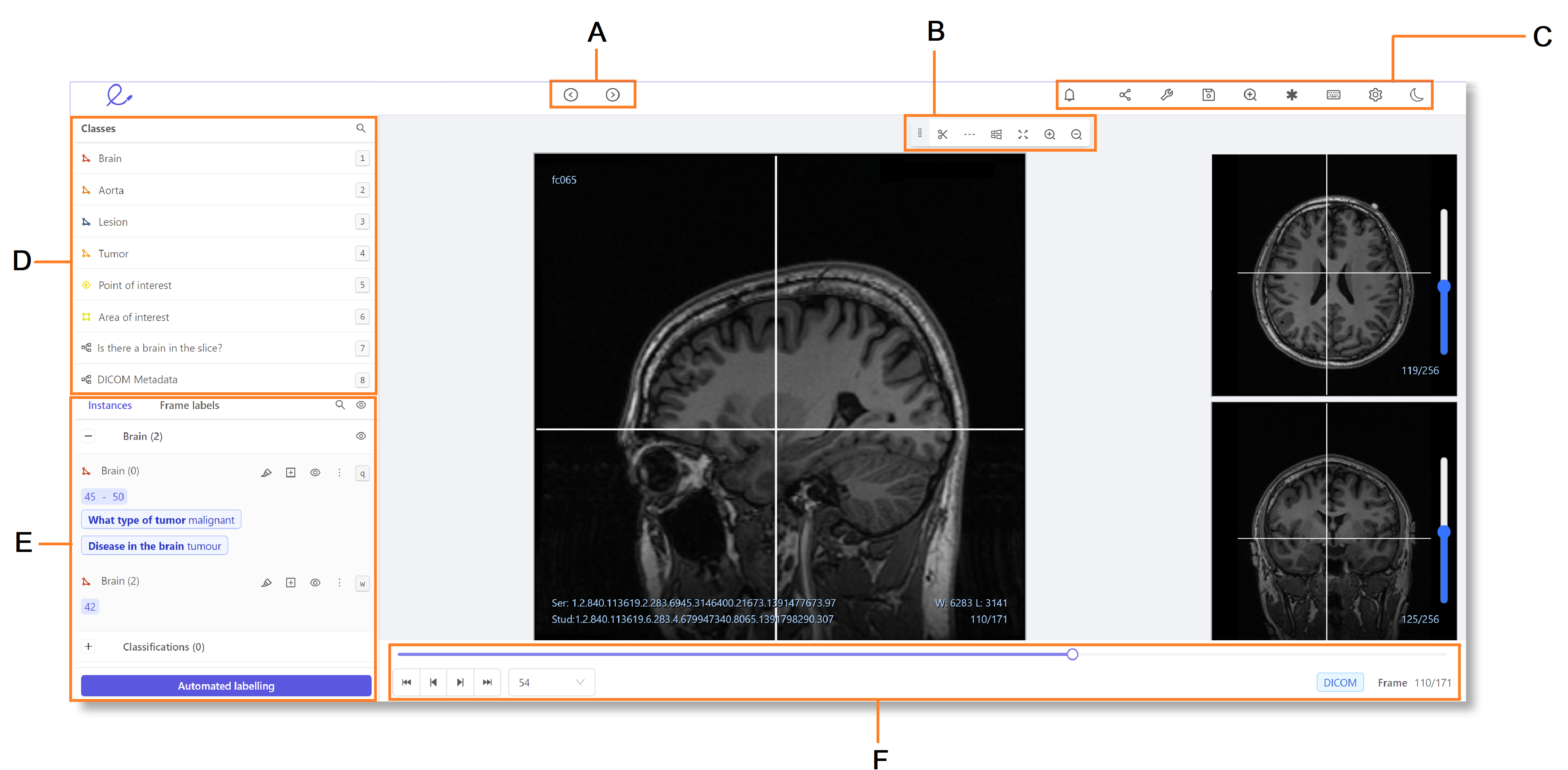
DICOM editor components:
- A. Load previous/next DICOM file (if viewing DICOM file outside the task manager)
- B. Toolbar with scissors (bulk vertex removal), measurement tools, windowing adjustment, image centering, and zoom
- C. Editor menu
- D. Slice slider and sliceskip interval
- E. Instances and frame labels
- F. Classes
The following sections describe general functionality for all DICOM modalities. If you are looking for Mammography specific features on the Encord platform please have a look here The Encord platform is very flexible, if you have questions regarding a specific modality/datatype please reach out to us at support@encord.com.
Task Management
Before we get into the details of the label editor and making annotations, a quick note about scheduling annotation work. Annotation is done in the annotation phase of the task management system. In order to ensure annotation work is saved properly, it's crucial that each labeling task is properly assigned to an annotator, annotators always access the task through the Queue tab in the Task Management System, and only the assigned annotator works on any given task at any given time.
Navigation
There are a number of different ways to skip through slices in a DICOM file:
- Using Left and Right arrow keys on your keyboard.
- Using the forward and backward buttons on the editor.
- By scrolling using your mousewheel or trackpad.
- By scrubbing through, or clicking anywhere on the progress bar.
"Slices" and "Frames" are used interchangeably in our DICOM editor documentation.
Hold the Shift key and drag to pan, and move a DICOM slice around on the page. To zoom in or out of a slice either hold Shift and use the Up and Down keys, or hold Shift while scrolling using your laptop's trackpad.
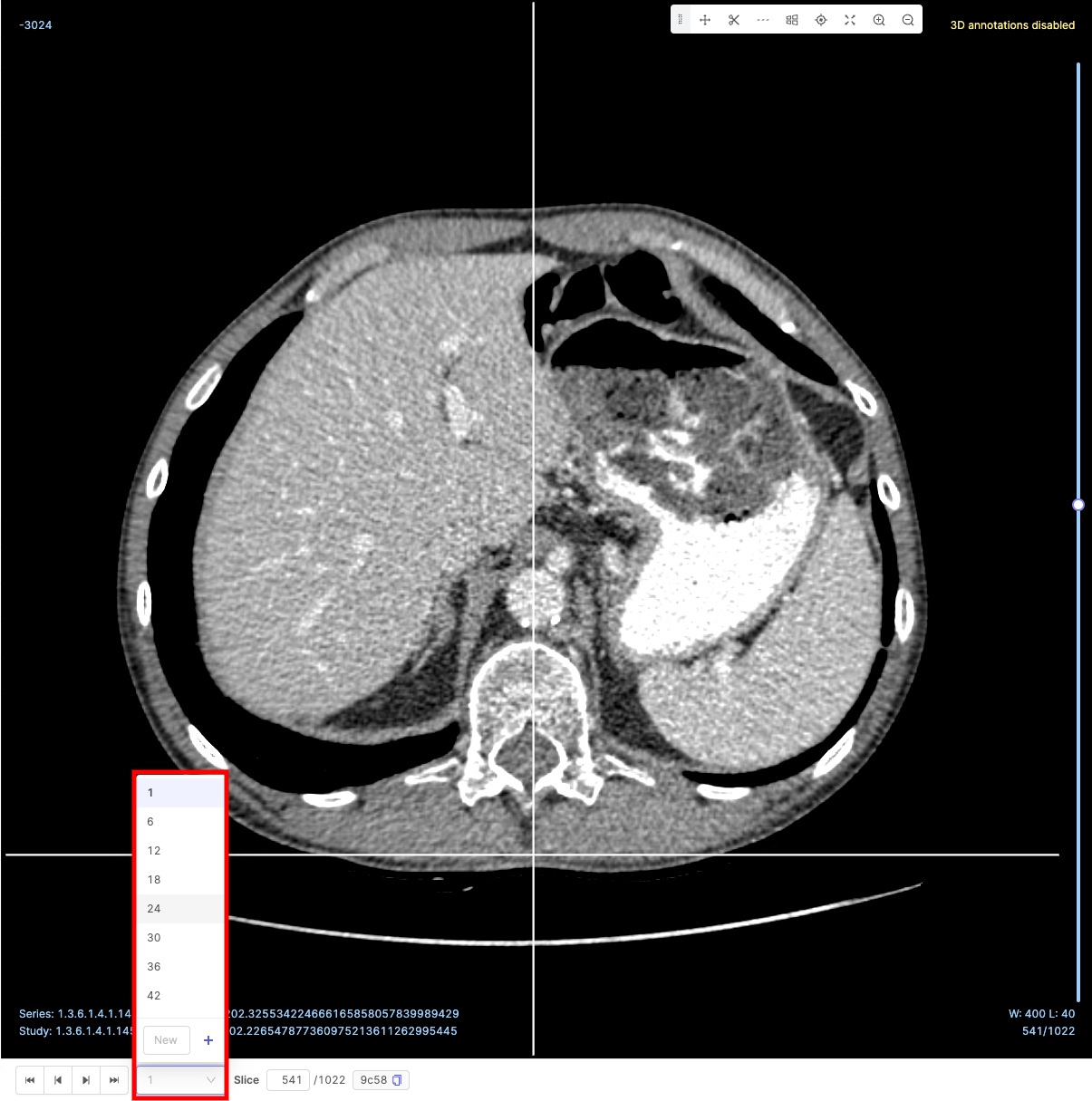
The number of slices skipped is determined by the specified Frameskip interval.
Frameskip intervals can be set using the predefined values, or customized by entering a value in the New input and clicking the ![]() icon.
icon.
Progressive loading
You don't have to wait for your file to finish loading before navigting to your desired frame! Simply drag the cursor to, or click the progress bar on, the location of the frame you would like to view while it is still loading, and the file will continue loading from the desired location, as seen in the video below. The light-blue color on the progress bar indicates how much of your file has been loaded.
Crosshair navigation
Crosshair mode can be activated using the ![]() icon in the floating bar.
Left clicking in a view will update the other views automatically so that the displayed image planes match the selected location.
icon in the floating bar.
Left clicking in a view will update the other views automatically so that the displayed image planes match the selected location.
Deeplinking - Copy and share URLs
Seamlessly copy the URL of the slice currently being viewed by clicking the ![]() icon, as shown below.
icon, as shown below.
Sharing a URL will now guide users to a specific image or slice within the project.
Display
Hounsfield Units
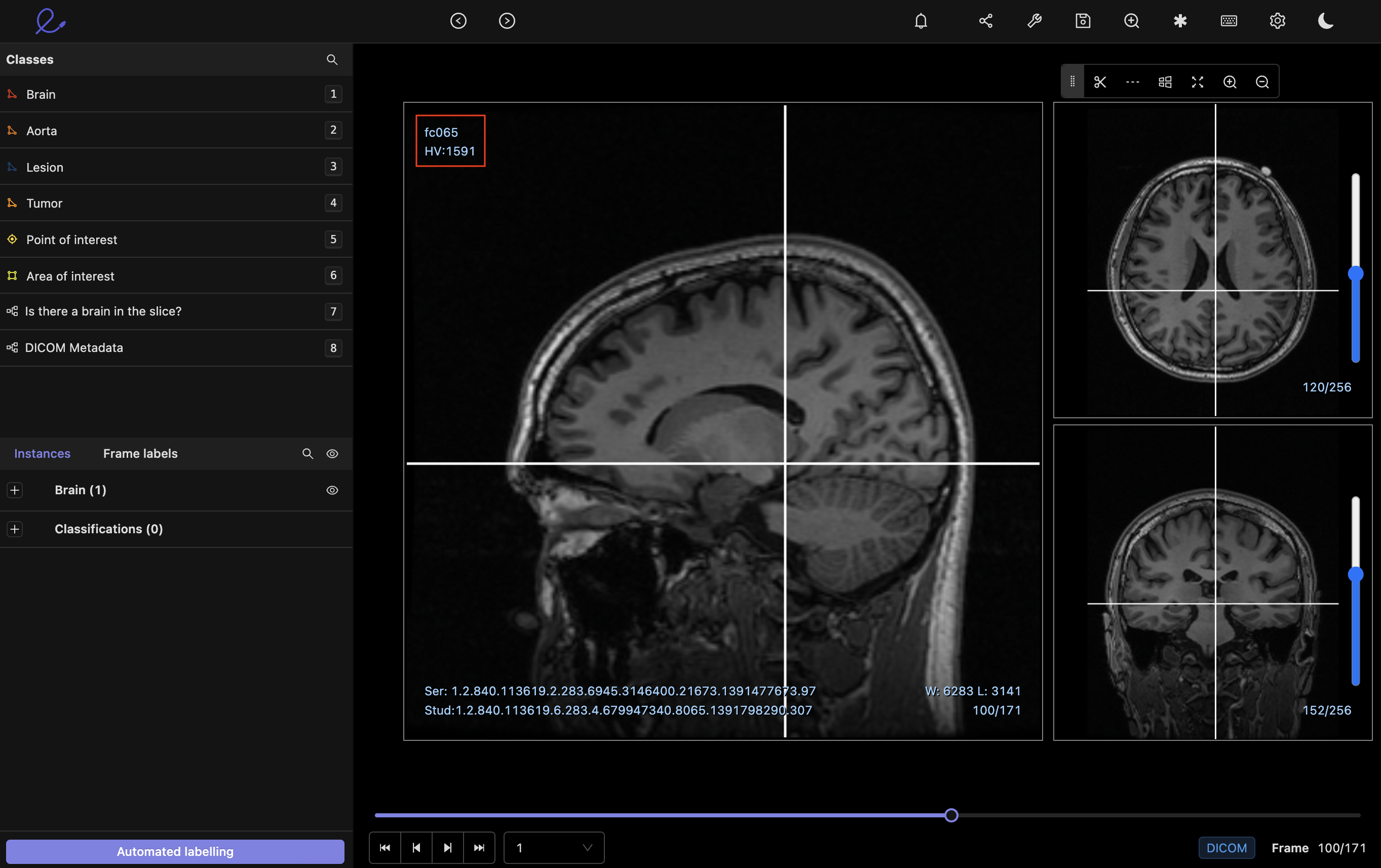
The Encord DICOM viewer renders DICOM files natively in the browser, which enables the display of the full range of the Hounsfield scale (i.e., 20,000+ pixel intensities). Hounsfield values are projected directly on the image. The example below has the cursor pointing to Hounsfield Value 1591.
2D Multiplanar reconstruction (MPR)
The MPR display provided within Encord reconstructs images in 2D orthogonal planes (coronal, sagittal, axial), helping to visualize, analyze, and annotate the anatomy, which is impossible with the base image alone. Annotations drawn on the main view will automatically show up in the corresponding orthogonal planes. Cross-reference lines are projected across the three views to aid you in accurate anatomy annotations.
The Encord DICOM viewer displays the base image in the left viewport for annotations, which can be either the coronal, axial, or sagittal view.
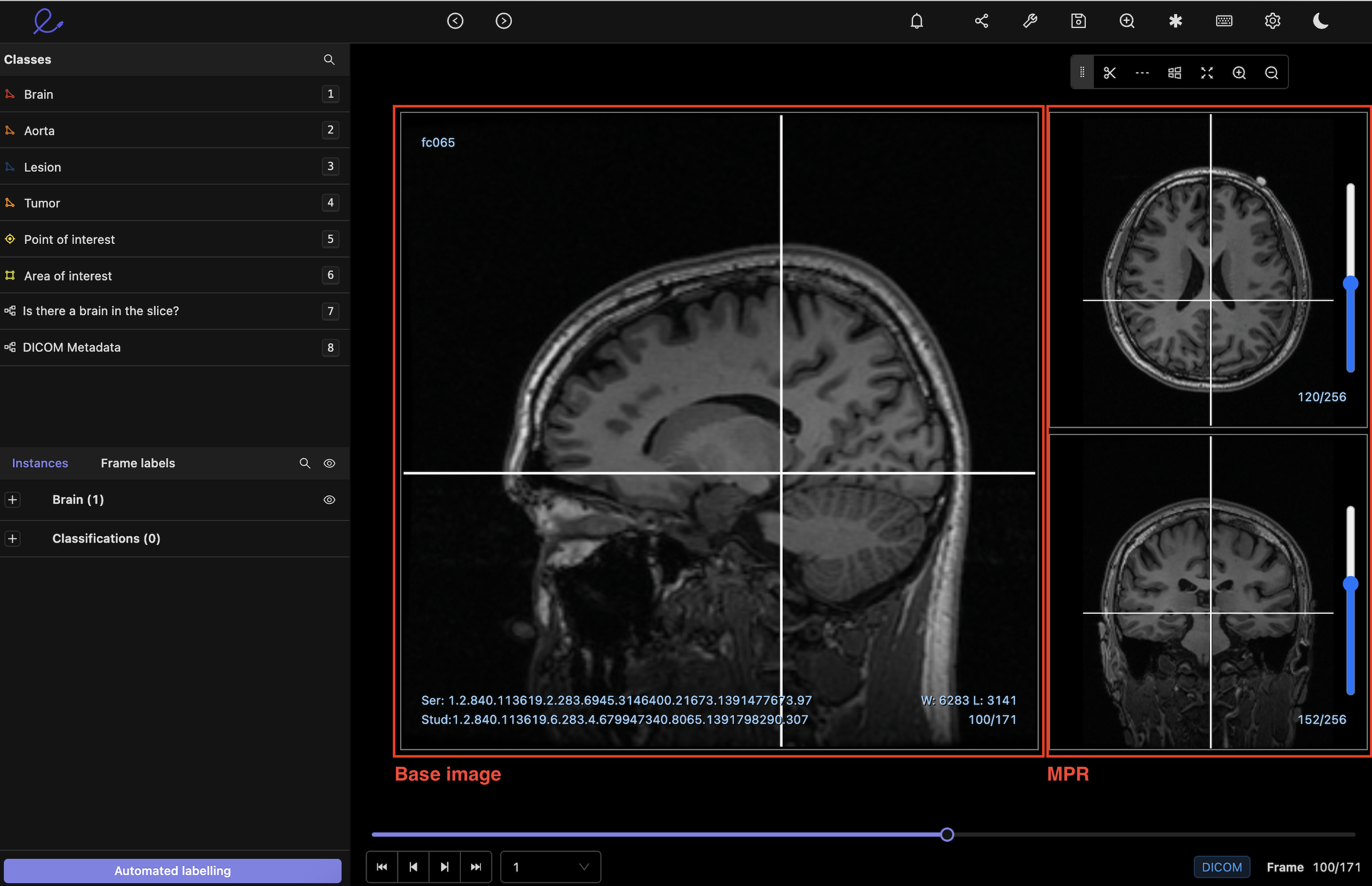
Our team is working on adding support for native 3D reconstruction.
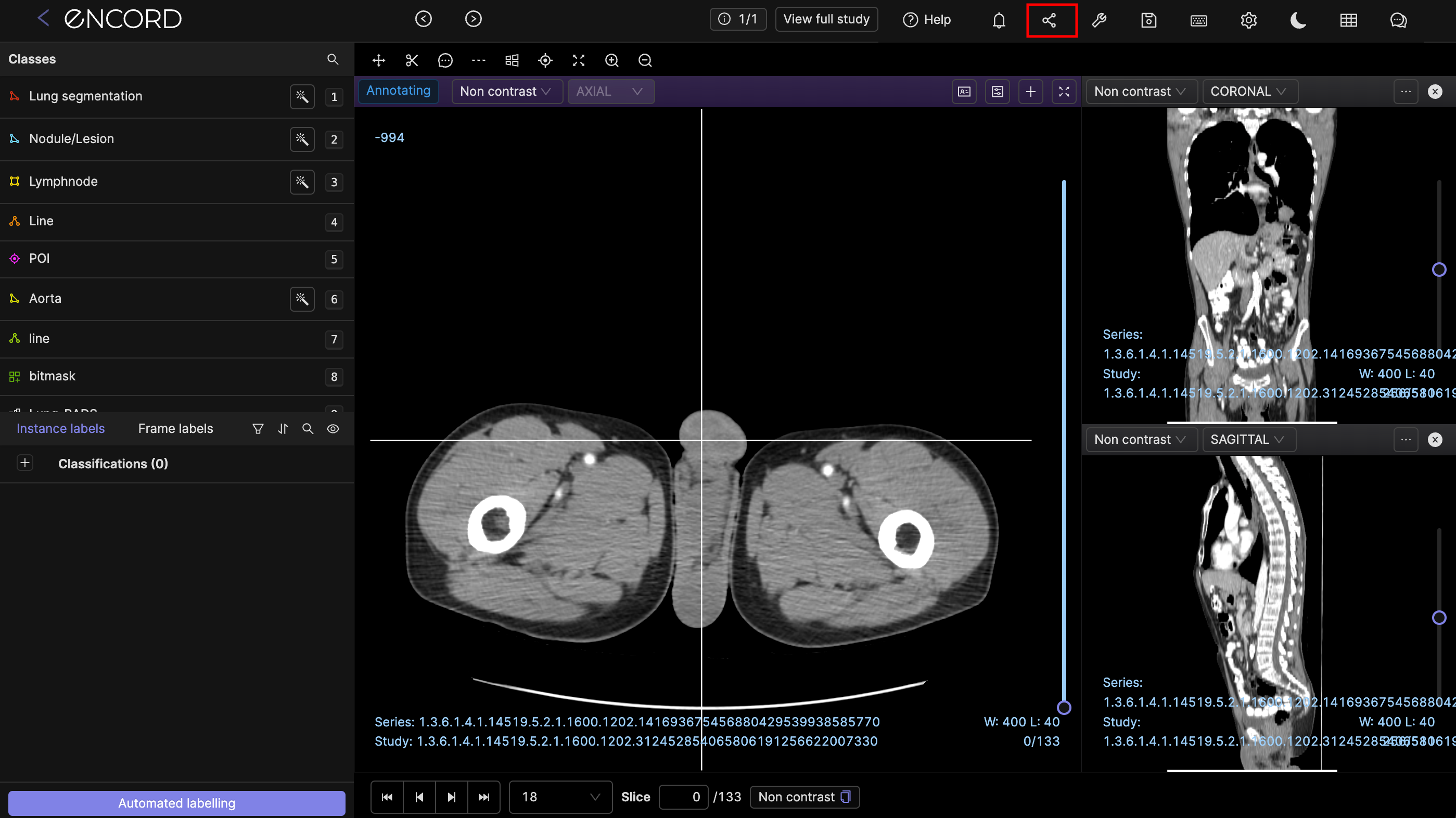
Multiple series viewing
In the dropdown menu of the viewport you can add other series to be viewed at the same time. Annotations will be seen on all the series being viewed, as seen in the screenshot below.
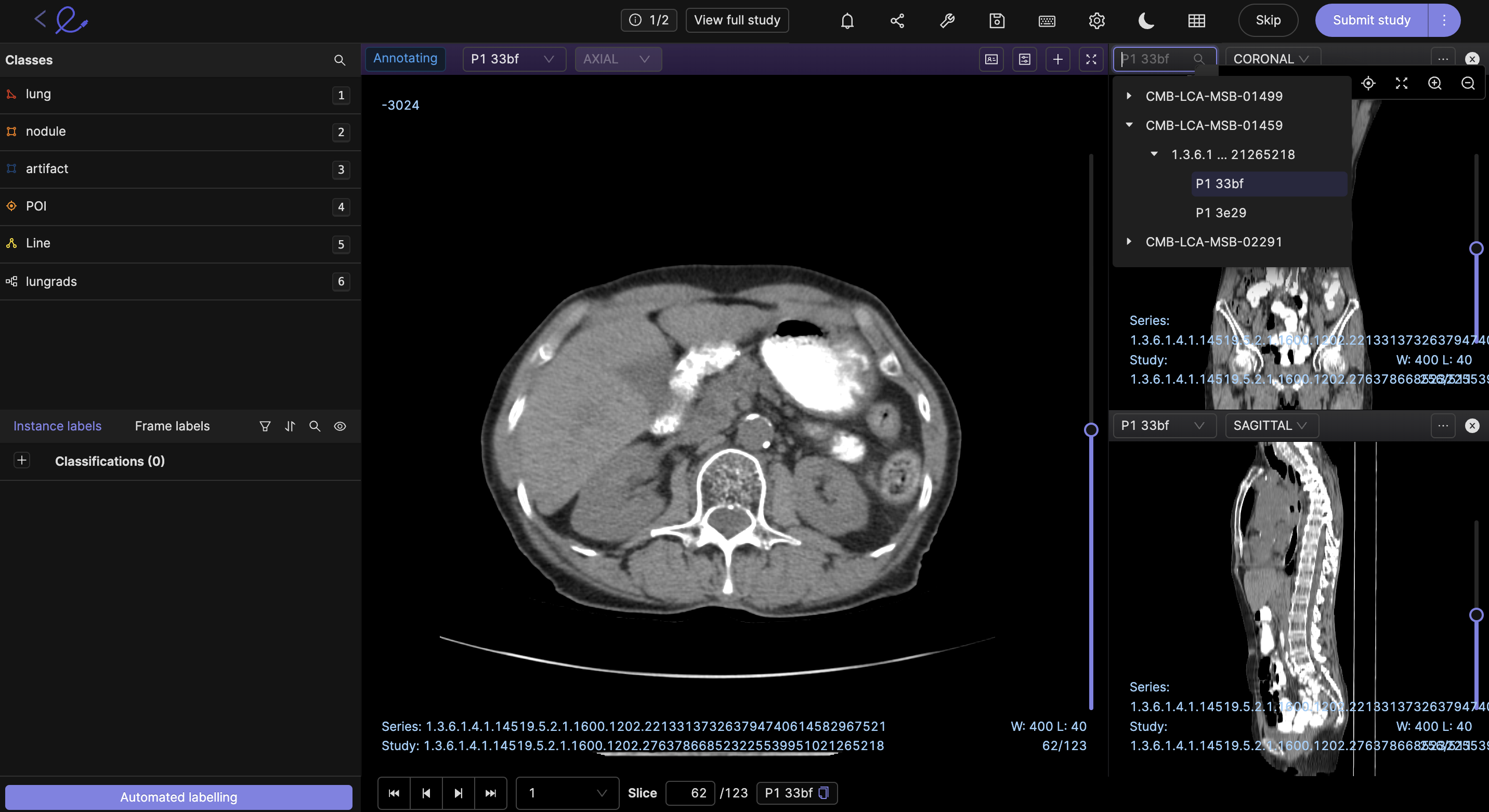
Only series that are part of the same dataset are available for selection. Thus, if you wish to view multiple series in one session, please ensure they're part of the same dataset.
We are currently working on a feature that will allow for synchronous scrolling of multiple series.
Window width (WW) and window level (WL)
Toggle the DICOM windowing settings menu by clicking the sliders icon in the viewer pane. Adjust window width and level using the sliders or setting desired input values. Save custom levels as presets for quick access later.
Use the more natural windowing adjustment functionality by clicking the Adjust windowing button in the toolbar or pressing alt + w on your keyboard. Next, press the left mouse button and drag the mouse to change the window width or level:
- Up to increase window level
- Down to decrease window level
- Left to shrink window width
- Right to expand window width
Maximum intensity projection (MIP)
Clicking the sliders icon in the corresponding viewer pane and then changing the slab thickness (ST) to something greater than 0 enables MIP mode. MIP can be switched on/off for each individual view. Note that annotations are not shown in MIP mode. To disable MIP mode change the ST back to 0.
Hanging protocols
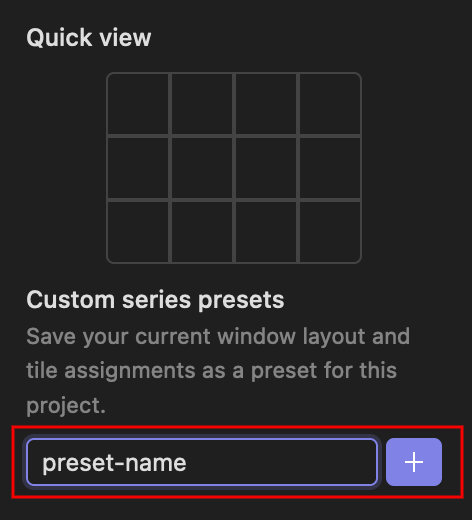
'Hanging protocols' allow you to select how many entities within a study are displayed at the same time. This allows you to, for example, view the same CT scan slice at different moments in time. For mammography files, the default setting is a 2x2 layout which allows you to view both breasts side-by-side and check for asymmetry between them.
Presets for a given case can be saved by simply typing the desired name after making a protocol selection, and clicking the + button.
Metadata
The Encord DICOM viewer renders DICOM files natively in the browser, allowing you to access metadata in the DICOM file.
Metadata viewer
The 'Metadata viewer' allows you to access all metadata pertaining to the series currently being annotated.
It is displayed by toggling the small ![]() button above the series being annotated.
The 'Metadata Viewer' contains a search bar allowing you to search for specific types of metadata.
button above the series being annotated.
The 'Metadata Viewer' contains a search bar allowing you to search for specific types of metadata.
A note will appear warning the user if 'Pixel spacing' is missing from an entity's metadata. Pixel spacing is essential for making accurate distance measurements.
Data overlay toggle
The 'Data overlay' toggle, located in the 'Additional controls' menu, allows you to display or hide metadata overlay. When toggled on, metadata is displayed in the bottom left corner of the series being annotated.
Custom DICOM metadata such as text reports from radiological descriptions can be rendered in popout drawers. Please email support@encord.com to add drawers with custom DICOM metadata.
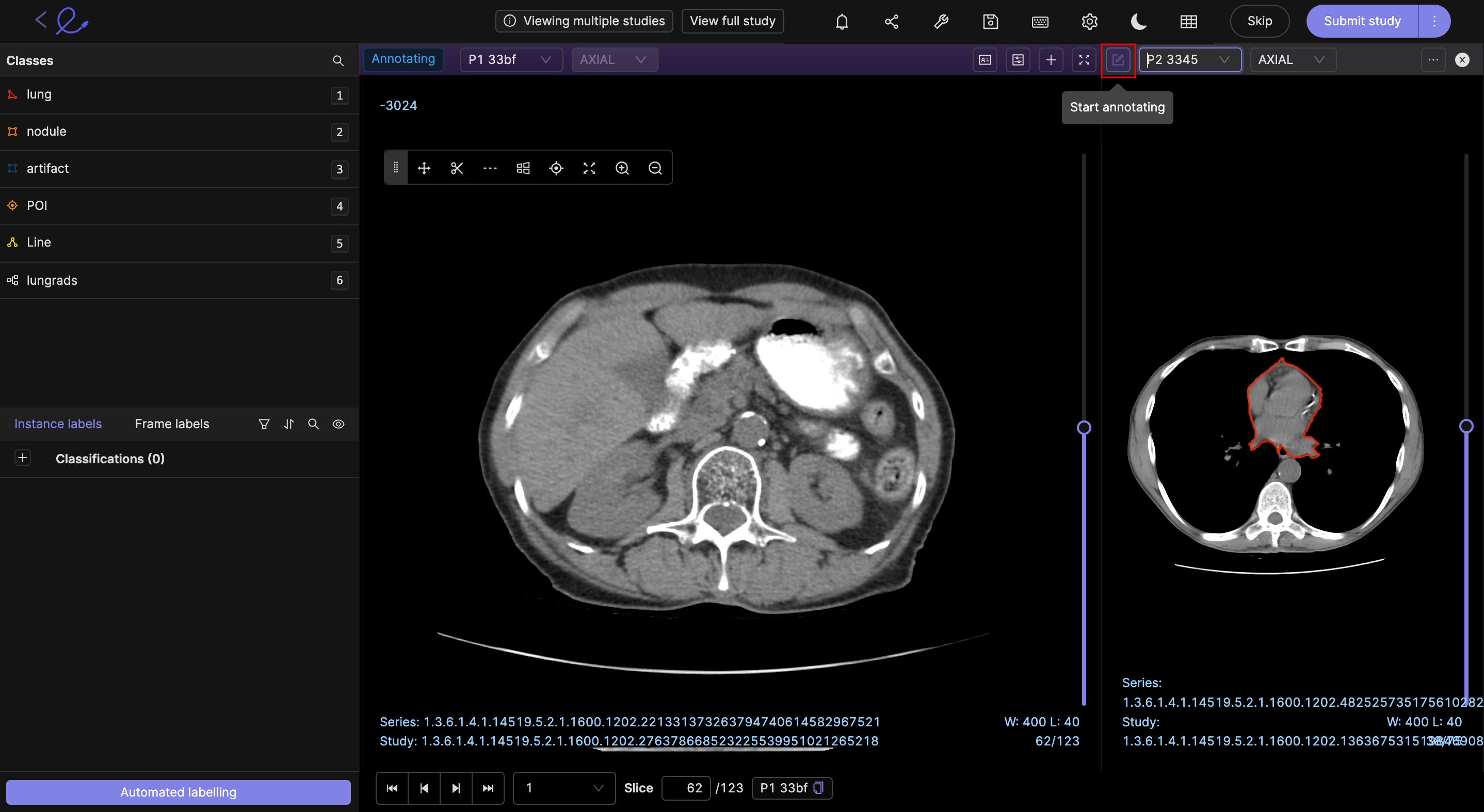
Editor switch
The 'Editor switch' button ( ![]() )
enables you to select any series being displayed to be annotated without having to return to the task management system.
)
enables you to select any series being displayed to be annotated without having to return to the task management system.
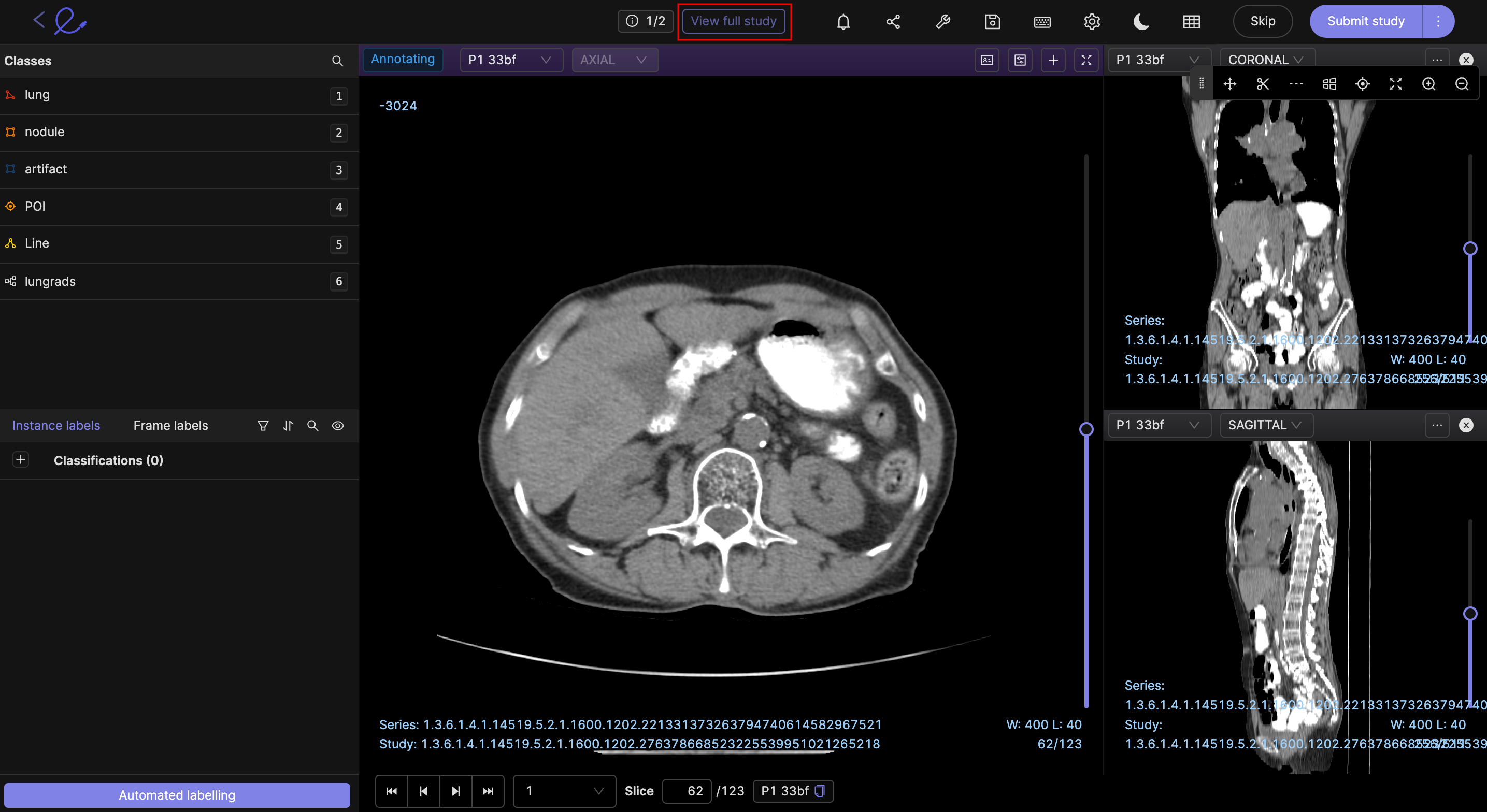
View full study
You can view the entire study which the series you are currently annotating is a part of, with a single click. The View full study button is highlighted in the image below.
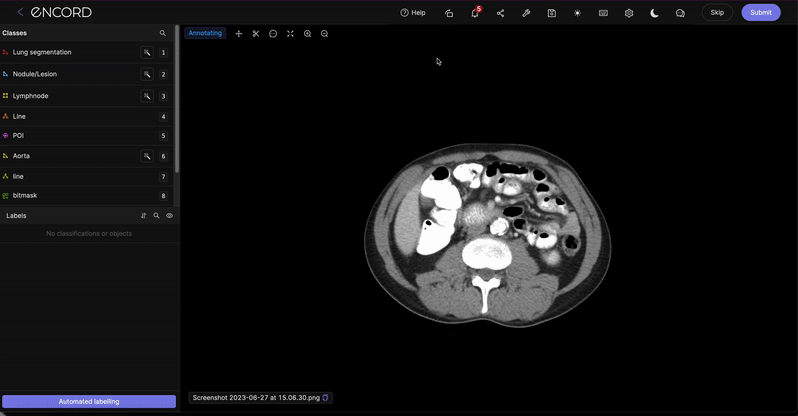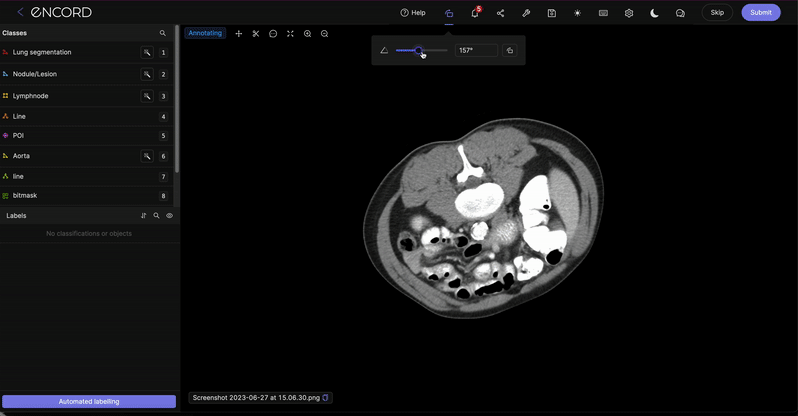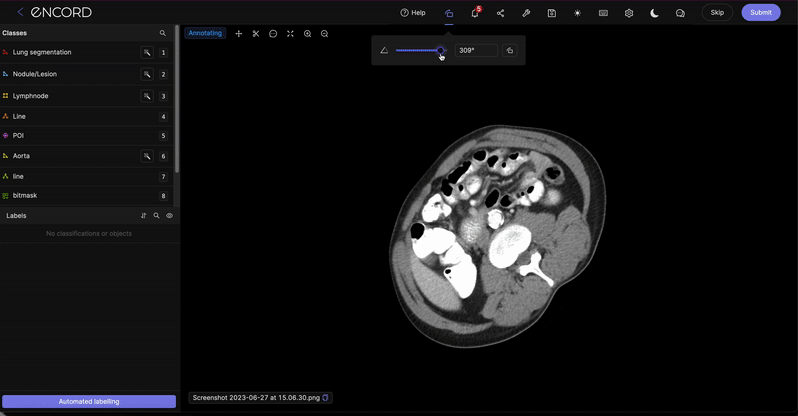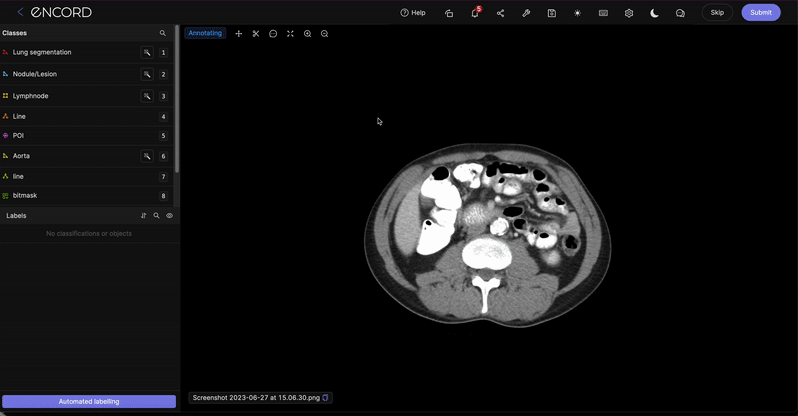
Rotate the label editor
Click the ![]() icon at the top of the label editor to bring up a pop-up enabling you to rotate the label editor by using the slider, as shown below.
icon at the top of the label editor to bring up a pop-up enabling you to rotate the label editor by using the slider, as shown below.
Click the ![]() icon to rotate the pop-up in increments of 90 degrees.
icon to rotate the pop-up in increments of 90 degrees.
Tools
Distance measurement
The distance measurement tool can be used to measure real-world distances between any two points on an image. It uses the 'Pixel spacing' parameter - contained in an entity's metadata - to accurately convert pixels into real-world distances that are displayed in millimeters.
For accurate measurements in mammography, the 'EstimatedRadiographicMagFactor' needs to be contained within an entity's metadata. It is used as a scale factor to adjust the distance measurement and provide accurate results. The absence of the 'EstimatedRadiographicMagFactor' can result in errors when measuring distances in mammography.
The Polyline ontology displays the total length of the line in millimeters, rather than simply the distance between two points,
Enable the distance measurement functionality by clicking the ![]() button on the floating toolbar, or by pressing alt + m on your keyboard.
button on the floating toolbar, or by pressing alt + m on your keyboard.
To delete existing measurements the distance measurement functionality has to be enabled. Erase measurements by selecting a measurement and hitting backspace or del on your keyboard.
Distance export
Encord supports copying measured distances using the polyline annotation type. Separate from using the measurement tool, as detailed above, create a polyline annotation as normal. Select the created polyline and click the three dots on the Quick toolbar to copy the distance to your clipboard.
Annotation types
Objects are annotated using bounding boxes, polygons, polylines, keypoints or primitives. Slices are annotated using classifications. Instantiating objects or classifications in the DICOM editor generates a UUID that uniquely identifies that instance across a range of slices (i.e., in the spatial dimension). The identifier is sometimes called a "track," with numerical IDs supplementing UUIDs for objects for visual aid.
Object instances can be assigned with static and dynamic classifications (i.e., spatial classifications that may change through the volume) as defined in the specified ontology. Static classifications define the global properties of an object (e.g., the surgical tool has three forks) whereas dynamic classifications change during the volume (e.g., the brain has a tumour present from slices 0 to 100 but none from frame 101 to 150).
Supported annotation types & corresponding features are listed in the table below. You can read more about the listed automation features here.
| Annotation type | Dynamic classification | Import predictions | Interpolation |
|---|---|---|---|
| Bounding box | ✅ | ✅ | ✅ |
| Polygon | ✅ | ✅ | ✅ |
| Polyline | ✅ | ✅ | ✅ |
| Point | ✅ | ✅ | ✅ |
| Primitive | ✅ | ✅ | ✅ |
| Frame classification | N/A | ✅ | N/A |
Bounding box
Creating a bounding box requires an ontology with a bounding box annotation type. Instantiate a new bounding box instance by clicking on the specified class in the 'Classes' menu, or by using the specified hotkey (e.g., 1, 2, 3).
Instantiating existing bounding box instances (i.e., bounding boxes that should keep the same identifier in preceding or succeeding slices) can be done by clicking on the Highlight icon for the specified object or using the assigned hotkey (e.g., q, w, e).
Bounding boxes can be copy/pasted between slices using cmd (MacOS)/ctrl (Windows) + c to copy the object to the clipboard and cmd/ctrl + v to paste it on the desired slice. See this visual illustration.
Bounding box instances can be assigned with static and dynamic classifications should they be defined in your ontology
- Highlight an object by clicking on it in the DICOM editor canvas or by clicking on the Plus icon or start/end slice in the range overview for the specified bounding box
- Assign a classifications by clicking on the relevant buttons or using the set hotkeys (e.g., q, u).
Interpolation
Interpolation is run via the 'Automated labeling' drawer. Click the Automated labeling button to open the drawer. Select *Interpolation from the dropdown menu, select the desired objects to interpolate, set the interpolation range (i.e., the range of slices) and click the Run interpolation button. Please note that you need a minimum two labels of an instance to run interpolation.
You can read more about interpolation here.
Polygon
Creating a polygon requires an ontology with a polygon annotation type. Enable or disable free-hand drawing mode by pressing d on your keyboard. Polygon coarseness for polygons drawn free-hand is set in the 'Settings' drawer in the Drawing settings dropdown. Instantiate a new polygon instance by clicking on the specified class in the 'Classes' menu or using the specified hotkey (e.g., 1, 2, 3).
A polygon can be closed by double-clicking anywhere on the canvas. Doing so will "snap" creating an edge between the last vertex to the first vertex drawn.
Instantiate existing polygon instances (i.e., polygons that should keep the same identifier in preceding or succeeding slices) by clicking on the Highlight icon for the specified object or using the assigned hotkey (e.g., q, w, e). See the keypoint documentation for visual instructions on how to instantiate existing polygons.
Polygons can be copy/pasted between slices using cmd (MacOS)/ctrl (Windows) + c to copy the object to the clipboard and cmd/ctrl + v to paste it on the desired slice.
Polygon instances can be assigned with static and dynamic classifications should they be defined in your ontology:
- Highlight an object by clicking on it in the editor canvas or by clicking on the Plus icon or start/end slice in the range overview for the specified polygon
- Assign a classifications by clicking on the relevant buttons or using the set hotkeys (e.g., q, u)
Interpolation
Run polygon interpolation following the instructions for bounding box, which you can find here. Unlike other linear interpolation methods, Encord's polygon interpolation algorithm does not require a matching number of vertices between polygon objects in set keyframes. Our algorithm also allows you to draw polygons in arbitrary directions (e.g., clockwise, counterclockwise, and otherwise).
You can read more about interpolation here.
Editing polygons with the brush tool
You can edit existing polygons with the brush tool. This allows you to create complex shapes in a much faster way than going vertex by vertex. To use the brush tool, select an existing polygon and then click on the brush tool button. This will show the brush tool settings panel where you can choose the brush or eraser and change its size. Once you are done editing, click the "x" to go back to the label editor.
Polyline
Creating a polyline requires an ontology with a polyline annotation type. Enable or disable free-hand drawing mode by pressing d on your keyboard. Polyline coarseness for polylines drawn free-hand is set in the 'Settings' drawer in the Drawing settings dropdown. Instantiate a new polyline instance by clicking on the specified class in the 'Classes' menu or using the specified hotkey (e.g., 1, 2, 3).
Polylines can be copy/pasted between slices using cmd (MacOS)/ctrl (Windows) + c to copy the object to the clipboard and cmd/ctrl + v to paste it on the desired slice.
Complete a polyline by double-clicking anywhere on the canvas.
Instantiate existing polyline instances (i.e., polylines that should keep the same identifier in preceding or succeeding slices) by clicking on the Highlight icon for the specified object or using the assigned hotkey (e.g., q, w, e).
Polyline instances can be assigned with static and dynamic classifications should they be defined in your ontology:
- Highlight an object by clicking on it in the editor canvas or by clicking on the Plus icon or start/end frame in the range overview for the specified polyline
- Assign a classification by clicking on the relevant buttons or using the set hotkeys (e.g., q, u)
Keypoint
Creating a keypoint requires an ontology with a keypoint annotation type. Instantiate a new keypoint instance by clicking on the specified class in the 'Classes' menu or using the specified hotkey (e.g., 1, 2, 3).
Instantiate existing keypoint instances (i.e., keypoints that should keep the same identifier in preceding or succeeding slices) by clicking on the Highlight icon for the specified object or using the assigned hotkey (e.g., q, w, e).
Keypoints can be copy/pasted between slices using cmd (MacOS)/ctrl (Windows) + c to copy the object to the clipboard and cmd/ctrl + v to paste it on the desired slice.
Keypoints instances can be assigned with static and dynamic classifications should they be defined in your ontology:
- Highlight an object by clicking on it in the editor canvas or by clicking on the Plus icon or start/end frame in the range overview for the specified keypoint
- Assign classifications by clicking on the relevant buttons or using the set hotkeys (e.g., q, u)
Interpolation
Run keypoint interpolation following the instructions for bounding box, which you can find here.
Primitives
Creating a primitive (f.k.a. skeleton templates) requires an ontology with a primitive annotation type. Use primitives to templatize shapes (example, 3D cuboids, pose estimation skeletons, rotated bounding boxes) commonly used by your annotation team.
To instantiate a new primitive instance, click on the specified class in the 'Classes' menu or use the specified hotkey (e.g., 1, 2, 3).
Instantiate existing primitive instances (i.e., primitives that should keep the same identifier in preceding or succeeding slices) by clicking on the Highlight icon for the specified object or using the assigned hotkey (e.g., q, w, e).
Primitives can be copy/pasted between slices using cmd (MacOS)/ctrl (Windows) + c to copy the object to the clipboard and cmd/ctrl + v to paste it on the desired slice.
Primitives allow you to define properties of edges defined in your template as visible, occluded, or invisible. Toggle the edge property settings for a primitive by highlighting the primitive and clicking the Show controls button.
Primitive instances can be assigned with static and dynamic classifications should they be defined in your ontology:
- Highlight an object by clicking on it in the editor canvas or by clicking on the Plus icon or start/end frame in the range overview for the specified primitive
- Assign classifications by clicking on the relevant buttons or using the set hotkeys (e.g., q, u)
Interpolation
Run primitive interpolation following the instructions for bounding box, which you can find here.
Segmentation masks / Bitmasks
Bitmasks allows you to create labels using a brush tool to select parts of an image. This can be useful when creating labels for vessel outlines, or labelling topologically seperate regions belonging to the same frame classification.
Creating a mask segmentation requires an ontology with the Bitmask annotation type.
To make changes to a label created using Bitmasks, the label has to be deleted and re-created.
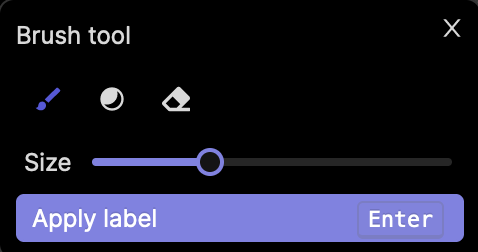
Brush tool
When you select your bitmask annotation type, the brush tool will be selected by default. You can adjust the size of the bitmask brush tool using the slider in the bitmask popup, as seen in the screenshot above.
The brush tool can be selected by clicking the ![]() icon, or by hitting f on your keyboard while the popup is open.
icon, or by hitting f on your keyboard while the popup is open.
When your label is ready, click Apply label, or press Enter.
Threshold brush
The Threshold brush tool specific to DICOM enables you to set an intensity value threshold for labels created using the bitmask annotation type. This means only parts of the image within the set intensity range will be labeled when selected using the Threshold brush tool.
To select the threshold brush, click the ![]() icon, or hit g on your keyboard while the popup is open. Adjust the size of the brush tool, as well as the range of intensity values you would like to include in your labels using the two sliders on the popup, as seen in the screenshot below.
icon, or hit g on your keyboard while the popup is open. Adjust the size of the brush tool, as well as the range of intensity values you would like to include in your labels using the two sliders on the popup, as seen in the screenshot below.
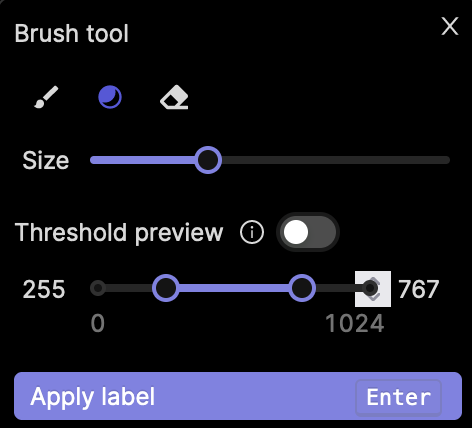
Use the Threshold preview toggle to see which parts of an image correspond to your set threshold. This allows you to see which parts of the image will be labeled when covered by the Threshold brush tool.
When your label is ready, click Apply label, or press Enter.
Eraser
The Eraser tool allows you to erase parts, or the entirety of your bitmask selection if the Apply label button has not been clicked yet.
To select the threshold brush, click the ![]() icon, or hit h on your keyboard while the popup is open.
icon, or hit h on your keyboard while the popup is open.
Frame classification
Creating frame (or slice) classification(s) requires an ontology with a classification annotation type. Frame-level classifications consider the frame as a whole, not the objects' localization. Instantiate a frame-level classification instance by clicking on the specified class in the 'Classes' menu or using the specified hotkey (e.g., 1, 2, 3).
Set the desired start and end frame by clicking the Set start to current and Set end to current buttons or using the set hotkeys c and v, respectively. Click the This frame button or the x hotkey to set the slider to only the current frame (e.g., frame 100). Add additional ranges by clicking the Add range button or pressing the n hotkey to add a frame range. Click the Save button or the m hotkey to save the instance. Use the This frame & save button or the z hotkey to classify and save the results for a single frame quickly.
Encord currently supports radio, checklist, and free-form text input classification types.
Edit existing classification instances by clicking on the Plus icon for the specified instance in the 'Classifications' pane in the 'All instances' section.
Dynamic classification
Static classifications define the global properties of an object (e.g., the surgical tool has three forks) whereas dynamic classifications change during the volume (e.g., the brain has a tumour present from slices 0 to 100 but none from frame 101 to 150). Assigning dynamic classification to objects requires an ontology with the dynamic classification flag toggled.
Encord supports dynamic classifications for all object annotation types (i.e., bounding box, polygon, polyline, keypoint, and primitives).
Highlight the desired object to assign dynamic classifications. Dynamic classifications are recognized by the Dynamic tag next to the given feature in the ontology. Dynamic classifications are only applied in the slices specifically indicated but can be edited in ranges if they span multiple slices. See the keypoint documentation for visual instructions on how to dynamically classify existing objects.
Edit the ranges of dynamic classifications by toggling the object menu dropdown, selecting the relevant dynamic classification, and clicking the Edit range button. Use the progress bar to change the specified range.
Automation
Interpolation
Encord's interpolation feature uses a proprietary linear interpolation algorithm that runs without using a representational model or matching pixel information in neighbouring frames.
Our interpolation algorithm has been built with pragmatic usage in mind. For example, unlike other linear interpolation methods, Encord's interpolation algorithm does not require a matching number of vertices between objects in set keyframes. Our algorithm also allows you to draw object vertices in arbitrary directions (e.g., clockwise, counterclockwise, and otherwise).
You can read more about interpolation here.
Auto-segmentation
Image segmentation refers to the process of dividing an image into multiple regions or sets of pixels, without constraints on the shape of said regions. The goal of an auto-segmentation tool is to tighten an original polygon segmentation to best match the edges of an enclosed object.
Please see our documentation on auto-segmentation to learn on to create, train, and use segmentation models.
Import model predictions
Encord's Python SDK & APIs allow you to import model predictions programmatically. Importing model predictions helps to pre-annotate your data to save annotation costs and use our quality control system for model validation.